
| AIUPred: Prediction of Intrinsically Unstructured Proteins |
 | DisCanVis: Integrating structural and functional annotations for the better understanding of the effect of cancer mutations located within disordered proteins. |
 | IUPred3: Prediction of protein disorder enhanced with unambiguous experimental annotation and visualization of evolutionary conservation |
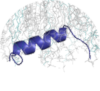 | MemMoRF is a comprehensive database of transmembrane and membrane associated proteins with MemMoRFs: regions that undergo disorder-to-order transition or large conformational changes upon membrane association. |
 | IUPred2A: Prediction of Intrinsically Unstructured Proteins |
 | PhaSePro: A comprehensive database of proteins driving liquid-liquid phase separation (LLPS) in living cells. |
Additional web servers and databases we are involved in
|
 | Disordered Binding Site (DIBS) database is a repository for protein complexes that are formed between Intrinsically Disordered Proteins (IDPs) and globular/ordered partner proteins. |
 | MobiDB: A database of protein disorder and mobility annotations |
 | DisProt: A database of intrisically disordered proteins. |